Paul Perco (1,2,3), Alexander Kainz (2), Gert Mayer (3), Arno Lukas (1,4), Rainer Oberbauer (2), and Bernd Mayer(1,4)
1 – Institute for Biomolecular Structural Chemistry, University of Vienna, Campus Vienna Biocenter 6, 1030 Vienna, AUSTRIA
2 – Department of Nephrology, Medical University of Vienna, Waehringer Guertel 18-20, 1090 Vienna, AUSTRIA
3 – Department of Nephrology, University of Innsbruck, Anichstrasse 35, 6020 Innsbruck, AUSTRIA
4 – emergentec biodevelopment, Rathausstrasse 5/3, 1010 Vienna, AUSTRIA
Implementation and Results
2. Biological data sets – 2.a. Actin beta (ACTB)
3 Promoter sequence sets
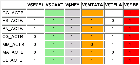
7 ACTB genes and 13 randomly picked genes.
View HTML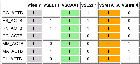
7 ACTB genes and 13 randomly picked genes.
View HTML
7 ACTB genes and 13 randomly picked genes.
View HTML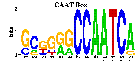
7 ACTB input sequences.
View HTML
7 ACTB input sequences.
View HTML
6 ACTB input sequences.
View HTML/////////////////////////////////////////////////////////////////////////////////////////////
2. Biological data sets – 2.b. Muscle-specific genes
JASPAR matrices used in the analysis // Regulatory regions which confer muscle-specific gene expression
Datasets after less stringent settings for MotifScanner
MotifScanner parameters: prior probability: 0.2; background model: EPD human-1st order
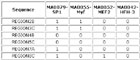
Input Matrix: 46 regulatory regions
View HTML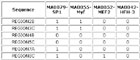
Input Matrix: 46 regulatory regions plus 40 random sequences
View HTMLDatasets after stringent settings for MotifScanner
MotifScanner parameters: prior probability: 0.1; background model: EPD human-3rd order
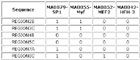
Input Matrix: 46 regulatory regions
View HTML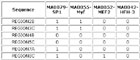
Input Matrix: 46 regulatory regions plus 40 random sequences
View HTML/////////////////////////////////////////////////////////////////////////////////////////////
3.Comparison to other routines – 3.a. Toucan ModuleSearcher (genetic algorithm approach)
Datasets after less stringent settings for MotifScanner
MotifScanner parameters: prior probability: 0.2; background model: EPD human-1st order
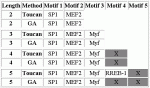
Modules detected: GA vs Toucan
View HTML
Modules detected: GA vs Toucan
View HTMLDatasets after stringent settings for MotifScanner
MotifScanner parameters: prior probability: 0.1; background model: EPD human-3rd order
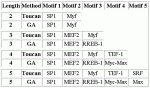
Modules detected: GA vs Toucan
View HTML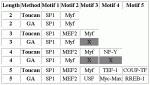
Modules detected: GA vs Toucan
View HTML
