Supplemental data: TRANSPLANTATION
Alexander Kainz(1), Paul Perco(1,2), Bernd Mayer(2,3), Afschin Soleiman(4), Rudolf Steininger(5), Gert Mayer(6), Christa Mitterbauer(1), Christoph Schwarz(1), Timothy Meyer(7), and Rainer Oberbauer(1)
1 – Department of Nephrology, KH Elisabethinen Linz, and Medical University of Vienna
2 – Institute of Theoretical Chemistry, University of Vienna
3 – emergentec biodevelopment GmbH, Vienna
4 – Department of Pathology, Medical Universtiy of Vienna
5 – Department of Transplant Surgery, Medical University of Vienna
6 – Department of Nephrology, Medical University of Innsbruck
7 – Department of Nephrology, Stanford University

Demographic data of kidney donors and recipients. Data are given as median and interquartile range.
BCAR … biopsy confirmed acute rejection
GFR … glomerular filtration rate estimated using the abbreviated MDRD formula
CIT … cold ischemia time
DM … diabetes mellitus
h_GFR … normal allograft function, calculated GFR > 45 ml/min/1.73 m2
l_GFR … higher impaired allograft function, calculated GFR < 45 ml/min/1.73 m2
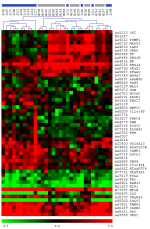
The dendrogram derived by unsupervised hierarchical clustering of differentially expressed genes between the groups l_GFR (calculated GFR < 45ml/min/1.73 m2) and h_GFR (calculated GFR ≥ 45ml/min/1.73 m2). The cosine correlation and complete linkage were used as distance measure and linkage rule in the hierarchical cluster algorithm, respectively. Red spots indicate abundantly expressed transcripts, whereas green spots indicate transcripts expressed on low level when compared to the reference RNA. GenBank accession numbers and Gene Symbols are used for labeling the cDNA clones.
View JPG
Representation of the differentially expressed genes between groups l_GFR (low GFR) and h_GFR (high GFR). Genes are categorized based on GO terms and PANTHER classifications and are ranked by difference of mean expression between the two groups under study. Values represent ratios of sample to standard reference expression on a log2 scale.
View PDF
Protein-protein interaction networks of up-regulated genes of group h_GFR (calculated GFR ≥ 45 ml/min/1.73 m2) and group l_GFR are given in figures 2A and 2B, respectively. Black boxes indicate differentially regulated proteins between the two groups; grey boxes represent proteins identified by the nearest neighbor expansion method. Proteins up-regulated in group l_GFR showed a high connectivity on the level of protein-protein interactions thus indicating concerted interaction. (C) The index of aggregation (IA; y-axis) in dependence of the number of proteins used for constructing protein interaction networks (x-axis). The IA of genes up-regulated in group l_GFR (calculated GFR < 45 ml/min/1.73 m2) is significantly above the IA of randomly selected lists. The thick black line represents mean IAs of randomly selected gene lists. Dotted lines represent two standard deviations of these distributions.
View JPG
RT-PCR validation of microarray experiments for BF, CR2, GRK5 and STAT1.
A. Retrospective validation.
View PDF
RT-PCR validation of microarray experiments for BF, CR2, GRK5 and STAT1.
B. Prospective validation.
View PDFOriginal data according to MiAME Guidelines
View PDFArray experiment:
l_GFR123 // l_GFR123_sher232.jpg // l_GFR123_sher232.gz
l_GFR138 // l_GFR138_sher240.jpg // l_GFR138_sher240.gz
l_GFR147 // l_GFR147_sher190.jpg // l_GFR147_sher190.gz
l_GFR162 // l_GFR162_sher228.jpg // l_GFR162_sher228.gz
l_GFR167 // l_GFR167_shem205.jpg // l_GFR167_shem205.gz
l_GFR190 // l_GFR190_sher209.jpg // l_GFR190_sher209.gz
l_GFR195 // l_GFR195_shem253.jpg // l_GFR195_shem253.gz
l_GFR206 // l_GFR206_sher182.jpg // l_GFR206_sher182.gz
l_GFR207 // l_GFR207_sher188.jpg // l_GFR207_sher188.gz
l_GFR216 // l_GFR216_sher238.jpg // l_GFR216_sher238.gz
l_GFR229 // l_GFR229_sher231.jpg // l_GFR229_sher231.gz
l_GFR232 // l_GFR232_sher221.jpg // l_GFR232_sher221.gz
l_GFR255 // l_GFR255_sher244.jpg // l_GFR255_sher244.gz
l_GFR258 // l_GFR258_sher245.jpg // l_GFR258_sher245.gz
l_GFR260 // l_GFR260_shem252.jpg // l_GFR260_shem252.gz
l_GFR262 // l_GFR262_shem254.jpg // l_GFR262_shem254.gz
h_GFR121 // h_GFR121_shem251.jpg // h_GFR121_shem251.gz
h_GFR130 // h_GFR130_sher180.jpg // h_GFR130_sher180.gz
h_GFR153 // h_GFR153_sher211.jpg // h_GFR153_sher211.gz
h_GFR155 // h_GFR155_sher189.jpg // h_GFR155_sher189.gz
h_GFR156 // h_GFR156_sher208.jpg // h_GFR156_sher208.gz
h_GFR160 // h_GFR160_sher210.jpg // h_GFR160_sher210.gz
h_GFR169 // h_GFR169_sher239.jpg // h_GFR169_sher239.gz
h_GFR172 // h_GFR172_sher198.jpg // h_GFR172_sher198.gz
h_GFR200 // h_GFR200_sher230.jpg // h_GFR200_sher230.gz
h_GFR201 // h_GFR201_shem206.jpg // h_GFR201_shem206.gz
h_GFR211 // h_GFR211_sher246.jpg // h_GFR211_sher246.gz
h_GFR213 // h_GFR213_sher222.jpg // h_GFR213_sher222.gz
h_GFR214 // h_GFR214_sher241.jpg // h_GFR214_sher241.gz
h_GFR227 // h_GFR227_sher242.jpg // h_GFR227_sher242.gz
h_GFR244 // h_GFR244_sher243.jpg // h_GFR244_sher243.gz

