P. Perco(1,2), A. Kainz(1), J. Wilflingseder(1), A. Soleiman(3), B. Mayer(2), R. Oberbauer(1,4)
1- Department of Nephrology, Medical University of Vienna & KH Elisabethinen, Linz, Austria
2- emergentec biodevelopment GmbH, Vienna, Austria
3- Department of Pathology, Medical University of Vienna, Austria
4- Austrian Dialysis and Transplant Registry, Austria

Demographic data of patients comparing the different CADI groups.
View PDF
Distribution of histological parameters. ati … acute tubular injury; ta … tubular atrophy; ii … interstitial inflammation; if … interstitial fibrosis; as … arteriolosclerosis; gs … glomerulosclerosis.
View PDF
Significantly enriched biological processes in upregulated DEGs are provided for the six histological parameters. The number in parenthesis depicts the number of DEGs that could be assigned to at least one category in the respective group. The number of genes for each category and histological parameter is provided where significantly enriched categories are indicated with an asterisk.
View PDF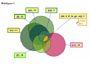
Distribution of histological parameters. ati … acute tubular injury; ta … tubular atrophy; ii … interstitial inflammation; if … interstitial fibrosis; as … arteriolosclerosis; gs … glomerulosclerosis.
View PDF
Linear regression model of histological and clinical parameters to predict creatinine values of one year after engraftment.
View PDF
Interaction network of DEGs and interacting proteins. DEGs are represented as grey filled nodes whereas interacting proteins connecting at least two DEGs are represented as smaller white nodes. Node border colors of DEGs depict the cluster assignment based on gene expression patterns. Node size of DEGs corresponds to the number of significant findings associated with the six histological parameters.
View PDF
Linear regression using molecular biomarkers to predict creatinine values of one year after engraftment.
View PDF
Scatterplot of predicted creatinine values to measured creatinine values after leave-one-out cross validation. The Pearson correlation coefficient between predicted and measured creatinine values was 0.45.
View PDF
Linear regression model of histological parameters represented as the CADI score to predict creatinine values of one year after engraftment.
View PDF
Model selection process with the seven molecular markers as independent variables to predict creatinine values one year after engraftment.
View PDF
Linear regression using molecular biomarkers in combination with clinical parameters to predict creatinine values of one year after engraftment.
View PDF
Original data according to MiAME Guidelines
View PDFData of all arrays in a single rar-File: Arraydata.rar (6.6 MB)

