Medical University of Vienna
>
AllFam
>
Help/FAQ
AllFam Help
The AllFam database is a resource for classifying allergens into protein
families. The database merges allergen data from the WHO/
IUIS Allergen
Nomenclature Database, supplemented by data from
AllergenOnline, with protein
family definitions from the Pfam
database.
See About AllFam for information on the AllFam team and how to
cite AllFam. See References for a list of references related
to AllFam and the underlying databases.
Background
In the last two decades, hundreds of allergens have been identified, cloned and
sequenced. This wealth of data now enables us to classify allergens into families of
evolutionary related proteins that are defined by sequence and structural similarity.
Members of the same family often have similar physico-chemical characteristics and
biochemical functions. Usually, only members of the same protein family show
cross-reactivity on the IgE and T cell level. However, it has to be stressed that neither
are all members of an allergen-containing protein family allergenic, nor are all allergenic
members of a protein family cross-reactive.
Employing different methods, several groups have shown that most allergens belong to a
limited set of protein families thus disproving the assumption that every protein can
become an allergen (see References). Hence, the
classification of allergenic proteins based on biochemical and structural similarities
including the comparison of allergenic and non-allergenic members of a protein family will
lead to new insights into factors that contribute to allergenicity. In addition, these data
will provide the foundation for elucidating the structural basis of allergenic
cross-reactivity.
[top]
Construction of AllFam
Figure 1 shows an overview of the algorithm used in constructing the
Allfam database. Briefly, the procedure is as follows:
- Merging data from the IUIS Allergen Nomenclature database and AllergenOnline:
- Sequences from AllergenOnline are matched with corresponding IUIS-approved allergens
from the same species by using a sequence identity threshold of 50% (applying a higher
threshold for very short fragments to exclude biologically insignificant short
alignments).
- Comparison with Pfam family definitions:
- Single sequences representing each allergen are compared with all Hidden Markov Models
(HMMs) representing protein families in Pfam using the thresholds for significant hits
defined in Pfam.
- Translation of found Pfam domains into AllFam families:
- This process in described in more detail below (What is the
difference between Pfam and AllFam families?).
- BLAST search of unclassified sequences against already classified ones.
- Most allergen sequences not classified by searching against Pfam HMMs are short
fragments obtained by N-terminal sequencing or mass spectrometry. These sequences are
compared with a database of already classified sequences using blastp and assigned to
the AllFam family of the most similar hit if the alignment yields a significant score.
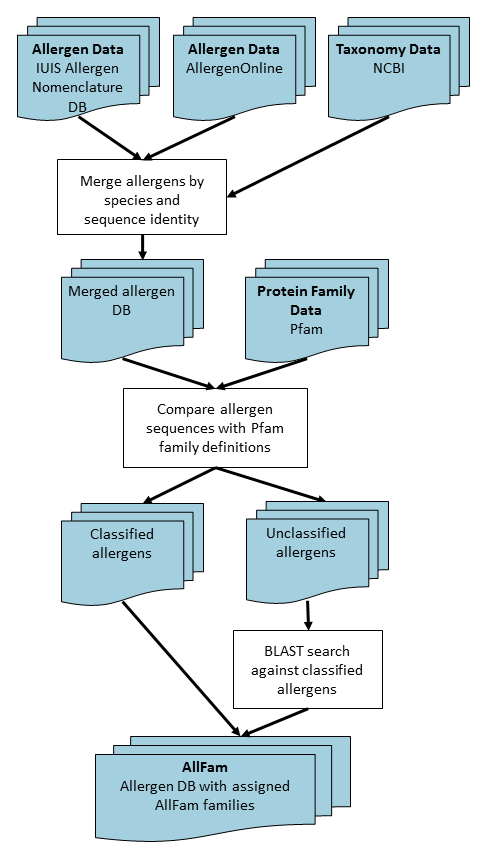
Fig. 1.
Flow chart showing the process of generating the AllFam database
[top]
What is the difference between Pfam and AllFam families?
The basis of AllFam's allergen family classification are protein family definitions from
Pfam. In contrast to Pfam domains, AllFam families are defined so that each allergen is
assigned to a single AllFam family and that related allergens are grouped into the same
AllFam family. This is achieved by the following steps:
- Merging of closely-related Pfam families into single AllFam families:
- Many families whose members show a low degree of sequence conservation cannot be
represented by a single Hidden Markov Model (HMM), but instead by a group of HMMs, each
of which represents only some family members. In AllFam, these groups of HMMs are merged
into single families. For instance, members of the EF-hand family
(AllFam:AF007) contain highly conserved calcium
binding sites flanked by helices with much lower degree of conservation. They are
represented in Pfam by multiple HMMs, six of which matched allergen sequences and, hence,
were merged into a single AllFam family.
- Merging of constituent domains of multi-domain proteins into single families:
- Some domains always occur together and are therefore merged into a single AllFam
family. For example, enolases consist of an N-terminal
(Pfam:PF03952) and a C-terminal
(Pfam:PF00113) domain, which are combined into a single
AllFam family: AF031.
- Classifying all constituent domains of different multi-domain proteins that share
a particular domain into a single family:
- Figure 2 shows the domain composition of members of AllFam family
AF043: Hevein-like and class I/II chitinase. The
family is defined by the hevein-like domain (Pfam:PF00187),
which is present in proteins with varying domain compositions. However, this AllFam family
also comprises class II chitinases, which lack a hevein domain. Otherwise, the chitinase
domain (Pfam:PF00182) would appear in two different AllFam
families.

Fig. 2.
Pfam domain compositions of members of AllFam family
AF043: Hevein-like and class I/II chitinase.
[top]
Search AllFam
You can search AllFam for various types of data:
- Allergen names
- IUIS-approved allergens are named by an abbreviation of the genus name (3-4 letters),
an abbreviation of the species name (1-2 letters) and a number, each item separated by a
blank (e.g. Der p 1 for the first allergen from
Dermatophagoides pteronyssinus). Non-IUIS allergens from AllergenOnline received
genus and species abbreviations according to the IUIS nomenclature, followed by a short
biochemical name or other unambiguous identifier (e.g. Hor v nsLTP-1 for the type 1
non-specific lipid transfer protein from Hordeum
vulgare).
- Allergen sources
- Search for allergen sources by species abbreviation, scientific name or common name.
For instance, search for peanut allergens by using "Ara h", "Arachis",
"Arachis hypogaea", or "peanut" as search terms.
- Family names
- Search for Allfam family names, names of the corresponding Pfam domains or keywords
associated with each family.
- AllFam family accession numbers
-
AllFam accession numbers have the format AFddd, i.e. the letters 'AF' followed by a
three-digit number.
- Pfam domain accession numbers
- Pfam accession numbers have the format PFddddd, i.e. the letters 'PF' followed by a
five-digit number.
All searches are case-insensitive, i.e. searches for 'bet v 1', 'Bet v 1' and 'BET V 1'
are equivalent. Multiple words are combined by AND, i.e. all words searched for have
to be found in order to yield a result.
[top]

